Walter Lab
The Walter Lab aims to identify principles of synaptic transmission as well as mechanisms by which synapses adapt their input/output relationship to stabilize signal transmission or process information.

In the Walter Lab, we aim to understand how the nervous system achieves its remarkable computational abilities. We study the molecular mechanisms of synaptic computation—synapses are not only the communication contact points in the nervous system but also the smallest computational units. Adapting communication across synapses forms the basis of all neural computation, including sensory processing, locomotion, learning, and memory. Our focus is on the fundamental mechanisms of how chemical transmitters are released from the cell that is sending information across the synapse and how the amount of transmitter is regulated under so-called synaptic plasticity.
To study these phenomena, we combine a wide array of approaches, integrating both experimental and theoretical research. We use Drosophila melanogaster as our model organism due to its simpler nervous system and synapses. Despite this simplicity, synaptic transmission is an evolutionarily well-conserved process, making our insights relevant across species. For instance, most known disease-related genes in humans are conserved in the fly. We employ electrophysiology, live and super-resolution microscopy, genetic and pharmacological manipulations to study how synapses adapt across all physiological timescales—from milliseconds to the lifetime of the organism. We also investigate the mechanisms of human disease by studying the functional consequences of human disease mutations on synapse and nervous system function in flies. Additionally, we build mathematical models to test hypotheses and predict outcomes for new experiments.
Our lab is funded by external, competitive research grants: In 2020, Alexander was awarded a Novo Nordisk Young Investigator Grant, and in 2022, an ERC Consolidator Grant. The lab currently consists of three postdoctoral researchers, a technical assistant, a PhD student, three research assistants, and a laboratory assistant.
- Jusyte, M., Blaum, N., Böhme, M.A., Berns, M.M.M., Bonard, A.E., Vamosi, A., Pushpalatha, K.V., Kobbersmed, J.R.L., and Walter, A.M. (2023)
Unc13A dynamically stabilizes vesicle priming at synaptic release sites for short-term facilitation and homeostatic potentiation.
Cell Reports https://doi.org/10.1016/j.celrep.2023.112541
Experimental and theoretical study of a novel presynaptic plasticity mechanisms regulating neurotransmitter release sites from milliseconds to minutes for temporal coding and homeostasis. - Kobbersmed, J.R.L, Berns, M.M.M, Ditlevsen, S., Sørensen, J.B., and Walter, A.M. (2022)
Allosteric stabilization of calcium and lipid binding engages several synaptotagmins in fast exocytosis
eLife https://doi.org/10.7554/eLife.74810
A theoretical work where we propose the first molecular model of Ca2+/Protein- and lipid interactions to achieve the high dynamic range for Ca2+-dependent neurotransmitter release. - Bohme, M. A., McCarthy, A. W., Blaum, N., Berezeckaja, M., Ponimaskine, K., Schwefel, D., and Walter, A. M. (2021)
Glial Synaptobrevin mediates peripheral nerve insulation, neural metabolic supply, and is required for motor function.
Glia; doi: https://doi.org/10.1002/glia.24000
Study on the glial/neural functional intersection. We show that SNARE-mediated secretion reactions in glial cells are required for metabolic supply, nerve function and proper animal locomotion. - Böhme, M. A., McCarthy, A. W., Grasskamp, A. T., Beuschel, C. B., Goel, P., Jusyte, M., Laber, D., Huang, S., Rey, U., Petzold, A., Lehmann, M., Göttfert, F., Haghighi, P., Hell, S. W., Owald, D., Dickman, D., Sigrist, S. J., and Walter, A.M. (2019)
Rapid active zone remodeling consolidates presynaptic potentiation.
Nat. Commun. 10, 1085, https://doi.org/10.1038/s41467-019-08977-6
This work establishes changes in synaptic ultrastructure as a mechanism for the consolidation of presynaptic potentiation in learning and homeostasis. - Reddy-Alla, S., Bohme, M. A., Reynolds, E., Beis, C., Grasskamp, A. T., Mampell, M. M., Maglione, M., Jusyte, M., Rey, U., Babikir, H., McCarthy, A. W., Quentin, C., Matkovic, T., Bergeron, D. D., Mushtaq, Z., Gottfert, F., Owald, D., Mielke, T., Hell, S. W., Sigrist, S. J., and Walter, A. M.# (2017)
Stable Positioning of Unc13 Restricts Synaptic Vesicle Fusion to Defined Release Sites to Promote Synchronous Neurotransmission.
Neuron 95, 1350-1364, https://doi.org/10.1016/j.neuron.2017.08.016
Bernard Katz's work in the 1950s established that neurotransmitters are released at only a few release sites, but their molecular identity was unknown for decades. By analyzing synaptic ultrastructure and physiology after genetic manipulation, my team was able to show that conserved Unc13 proteins define these release sites.
Personal Information
| Name: | Dr. rer. nat. habil. Alexander Matthias Walter, MSc |
| Date of birth: | 08.01.1981 |
| citizenship: | German, British |
| email: | awalter@sund.ku.dk |
| Website: | https://www.walter-lab.com/ |
| Orchid ID: | 0000-0001-5646-4750 |
| Google Scholar: | https://scholar.google.de/citations?user=4rT8Y30AAAAJ&hl=de# |
Positions Held
| 2020-now | Associate Professor for Molecular and Theoretical Neuroscience, Department of Neuroscience, Faculty of Health and Medical Sciences, Copenhagen University, Denmark |
| 2015-2021 | Emmy Noether Research Group Leader (part time since Aug. 2021), of the Group “Molecular and Theoretical Neuroscience” at the Leibniz-Forschungsinstitut für Molekulare Pharmakologie, Berlin, Germany |
| 2013-2015 | Postdoc, with Volker Haucke, Neurocure Cluster of Excellence, Charité Berlin, Germany |
| 2011-2013 | Postdoc with Jakob Sørensen, Department of Neuroscience and Pharmacology, University of Copenhagen, Denmark |
| 2010-2011 | Postdoc with Matthijs Verhage, Center of Neurogenomics and Cognitive Research, Vrije Universiteit Amsterdam, the Netherlands |
Education
| 2023 | “Habilitation” (venia legendi, qualification as a University Professor) in Physiology with Prof. Dr. Jörg Geiger, Charité University Hospital, Berlin, Germany |
| 2006-2010 | PhD with Jakob Sørensen, Erwin Neher, Reinhard Jahn, Department of Membrane Biophysics, Max-Planck Institute for Biophysical Chemistry, Göttingen, Germany, grade dissertation: summa cum laude |
| 2006 | Master, International Max-Planck Research School (IMPRS) and Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden – Note: A (excellent), top of the class |
| 2003 | Vordiplom, Chemistry, Georg August Universität Göttingen, Germany – grade: Sehr gut |
| 2000 | Abitur (German A-levels), Gymnasium Walsrode, Germany – grade: 1.3, top of the class |
3rd Party Funding
| 2022 | ERC consolidator grant (2 000 000 EUR), European Union, Brussels, Belgium |
| 2019 | Novo Nordisk Young Investigator Award (25 000 000 DKK, ~3 350 000 EUR), Award to conduct a research project at Copenhagen University, Denmark |
| 2016-now | SFB/TRR 186 Research Grant (471 120 EUR), project leader in the “Transregio“ TRR186 “Molecular Switches in the Spatio-Temporal Control of Cellular Signal Transmission“ between Heidelberg and Berlin funded by the “Deutsche Forschungsgemeinschaft” (DFG), Bonn, Germany |
| 2015-now | Emmy Noether Research Grant (2 051 700 EUR), Funding of research group, DFG Bonn, Germany |
| 2010-2011 | EMBO Long-term Fellowship (~35 000 EUR), Funding of a postdoc position |
Honours and Funding
| 2017 | Japan Neuroscience Society, Award for excellent presentation, 40th annual meeting, Tokyo, Japan |
| 2016-now | Einstein Center for Neuroscience Berlin (~35 000 EUR), funding of a PhD position |
| 2009 | 59th Meeting of Nobel Laureates, Invitation to the annual meeting of Nobel Laureates 2009 |
| 2008 | Excellence in Teaching Neuroscience Award, Award for the best teaching (tutorial) in the IMPRS Neuroscience curriculum, Max-Planck Research School, Göttingen |
| 2007 | Symposium Award, Award by the Society of General Physiologists, Woods Hole, MA, USA |
| 2005-2006 | Fellowship “Deutscher Akademischen Austauschdienst“ (DAAD) for a Master’s Thesis in Sweden (Karolinska Institute Stockholm), Bonn, Germany |
| 2004-2005 | Fellowship Max-Planck Society, Max-Planck Research School Neuroscience, Göttingen, Germany |
Teaching Experience
| since 2022 | Lectures and practical exercises in Physiology, Curricula for medical students and dentistry students, University of Copenhagen, Denmark |
| since 2021 | Lecture & Seminars in Neuroscience I: Cells and circuits. University of Copenhagen, Denmark |
| 2018 | FENS Cajal course: Advanced imaging methods for Cellular Neuroscience, Instructor, Bordeaux, France |
| since 2018 | Seminars in Neurophysiology, Curriculum for medical students, Charité Berlin, Germany |
| since 2017 | Lecture: Neurophysiology for Dentistry Students, Charité Berlin, Germany |
| 2014 | Lecture and practical course in the Master’s Modul “Molecular Neurogenomics”, Freie Universität Berlin, Germany |
| 2010 | Coordination of course “Advanced Neurogenomics“, Vrije Universiteit Amsterdam, Netherlands |
| 2006-2007 | Tutor to Erwin Neher’s Lecture „Electrophysiological Techniques“, Max-Planck Research School, Göttingen, Germany |
Didactic qualilfications
| 2021 | Qualification as German University Professor “Habilitation” with Prof. Dr. Jörg Geiger, director of the Institute for Neurophysiology, Charité University Hospital, Berlin, Germany |
| 2020 | Didactically certified implementation of a teaching project: Conception of a lecture series in physiology, assessed by the “Dieter Scheffner Fachzentrum” for University didactics, Charité University Hospital, Berlin, Germany |
| 2020 | Certified didactic training (40 hours), “Dieter Scheffner Fachzentrum” for University didactics, Charité University Hospital, Berlin, Germany |
| 2017 | Certified didactic training (21 hours), Centre for Science & Research Management, Speyer, Germany |
| 2008 | Excellence in Teaching Neuroscience Award, Award for the best teaching (tutorial) in the IMPRS Neuroscience curriculum, Max-Planck Research School, Göttingen |
Invitation as Speaker to international conferences
| 2024 | EU Synapse Meeting, Berlin, Germany, Talk: “Unc13A dynamically stabilizes vesicle priming at release sites for short-term facilitation and homeostatic potentiation” |
| 2023 | Meeting of the German Physiological Society, Berlin, Germany, Talk: “Molecular mechanisms of fast and sustained presynaptic potentiation” |
| 2022 | Neurofly conference, St Malo, France, Talk: “Neurotransmitter release sites adapt on millisecond and minute timescales for presynaptic plasticity” |
| 2020 | Baeza meeting of the SynGo Consortium, Baeza, Spain, Invitation to the workshop: “Synaptic Dimension of Brain Disorders Workshop” –postponed to 2021 due to Covid-19 |
| 2019 | Biophysical Society Thematic Meeting, Padova, Italy, Symposium talk: “Release site recruitment and activation as mechanisms of presynaptic plasticity” |
| 2019 | Dutch Neuroscience Meeting, Lunteren, the Netherlands, Key note lecture: “Release site addition as a mode of presynaptic plasticity” |
| 2018 | Federation of European Neuroscience Societies (FENS)-forum, Berlin, course “Introduction to synaptic transmission at the central synapse”, Talk: “Synapse structure/function and diversity” |
| 2018 | EMBO Workshop “Exocytosis and Endocytosis”, Tenerife, Spain, Symposium talk: “Unc13A generates and positions synaptic vesicle release sites” |
| 2017 | International Symposia of Chromaffin Cell Biology, Sheffield, United Kingdom, Symposium talk: “Phosphatidylinositol 4,5-bisphosphate optical uncaging potentiates exocytosis in chromaffin cells” |
| 2017 | 40th Annual Meeting, Japanese Neuroscience Society, Tokyo, Japan, Symposium talk: “Tight distribution of synaptic vesicle release sites generated by Unc13 synchronizes neurotransmission” |
| 2014 | INCF Neuroinformatics Congress, Leiden, the Netherlands, Symposium talk: “A catalytic slot model for exocytosis with a single release sensor effectively explains properties of neurosecretion” |
| 2011 | Exocytosis and Endocytosis Meeting, Edinburgh, United Kingdom, Talk: „A sequential secretion model with a single Ca2+-sensor explains heterogeneity in release kinetics” |
Organisation of Scientific Meetings
| 2025 | Synapse Physiology in Health in Disease, Copenhagen, Denmark, Symposium Organizer |
| 2024 | 4th Nordic Neuroscience Meeting, Session chair “New Frontiers in Synapse Biology”. |
| 2024 | Core2core Symposium Göttingen, Germany, Symposium Co-organizer “Synaptic transmission”. |
| 2016 | Federation of European Neuroscience Societies (FENS)-forum, Copenhagen, Denmark, Chair of Symposium S26 “Modelling of the synapse- the presynaptic side”. |
Current Collaborators
|
Carlotta Martelli, Gutenberg University Mainz, Germany. Investigating the relevance of presynaptic plasticity for sensory adaptation |
|
Stefanie Winkelmann, Zuse Institute Berlin, Mathematical modelling of synaptic transmission |
|
Shigeki Watanabe, Johns Hopkins University, Baltimore, US. Molecular analysis of transient vesicle docking and synaptic plasticity. |
| Dion Dickman, Department of Biology, University of Southern California, Los Angeles, USA. Physiology of synaptic transmission |
| Susanne Ditlevsen, Department of Mathematical Sciences, University of Copenhagen, Denmark. Mathematical modelling of synaptic transmission |
| Stephan Hell, Department for NanoBioPhotonics, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany. Super-resolution microscopy |
| André Nadler, Max Planck Institute for Molecular Cell Biology and Genetics, Dresden, Germany. Investigation of signalling lipids using “lipid uncaging” |
| David Owald, Institute for Neurophysiology, Charité Berlin, Germany. Behavioural Analyses (learning and memory) of Drosophilae |
| Köksal Özgül, Haccettepe University, Ankara, Turkey. Investigation of synapse function in Drosophila models of human neurometabolic diseases. |
| Christian Rosenmund, Institut für Neurophysiologie, Charité Berlin. Molecular mechanisms of exocytosis. |
| David Schwefel, Institut for Medical Physics and Biophysics, Charité Berlin. Structural Biology |
| Stephan Sigrist, Institute for Genetics, Freie Universität Berlin, Germany. Drosophila Neurogenomics and synaptic ultra-structure |
| Matthijs Verhage, Vrije Universiteit Amsterdam, The Netherlands. Neural disease gene ontology. |
Fly genetics
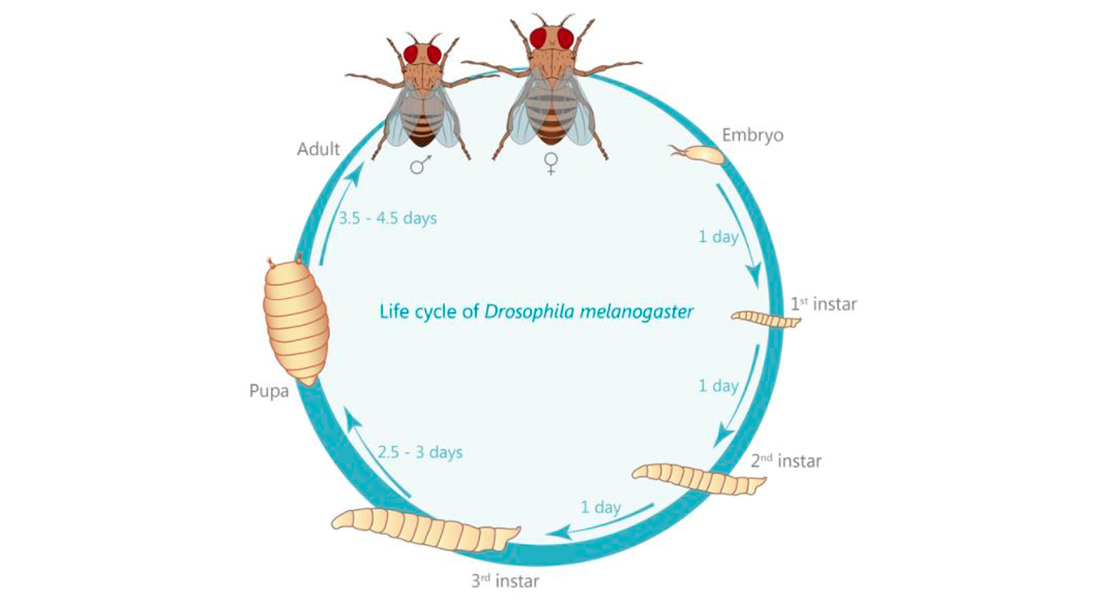
The fruit fly Drosophila melanogaster is very amenable to genetic modification. From the adult fly laying eggs to the 3rd instar larva we’re preferentially working with, it only takes 7-9 days. Using genetic toolboxes developed by other scientists, we can tag our protein of interest with fluorescent markers, or remove a protein the role of which we want to study.
Since its initial use by William Castles and Thomas Hunt Morgan more than 100 years ago, Drosophila melanogaster has become one of the most heavily used model organisms to study a diverse range of biological processes including genetics, embryonic development, learning, aging, and neurobiological processes. Its initial use by Morgan redefined the theory of heredity by discovering genes and found that they are located within chromosomes. This led to the first of six Nobel prizes awarded for Drosophila research (the others were awarded for the discovery that mutations are caused by X-ray, for founding the field of evolutionary developmental biology, and for the discovery of olfactory receptors, the innate immunity and finally of the circadian rhythm).
Interestingly, one convention unique to Drosophila was introduced by Morgan: the naming of genes in accordance to the phenotype their knock-down causes: the first mutation ever isolated is known as white, causing white instead of the wild type red eyes. Over the years, this convention has also led to somewhat entertaining names as “cheap-date”, naming flies that suffer from a reduced alcohol tolerance; or the opposite situation “happy hour”, naming flies that show an increased tolerance (Corl et al., 2009; Moore et al., 1998). For the research questions our lab is focused on, Drosophila is unique in its advantages. These include a relatively short generation time of about 10 days (see life cycle), an every growing genetic tool box allowing tissue- and even cell-specific expression or knock-down of genes of interest but especially a tremendous applicability to scientific methodologies (see methods section). Furthermore, 75% of all known human disease related genes have a recognizable counterpart in the Drosophila genome (Jennings, 2011) making it a perfectly suited tool to not just understand the molecular underpinnings of humans diseases, but also to study the function of these molecules in the healthy organism.
Live imaging
Biological processes are best viewed in real time and with as much temporal and spatial information as possible. Therefore, we use fluorescent probes to visualize synaptic activity and protein trafficking in live dissected larvae.
The fluorescent calcium indicator GCaMP has proven especially useful in this, and we use it to watch as the synapse releases its vesicles. Using this method, we measure the influence of synaptic proteins on signal transmission which helps us understand how the synapse works.
Super-resolution microscopy: STED
A large extent of the current understanding of biological processes comes from experiments that visualized them using various microscopic techniques including fluorescence microscopy. However, due to the Abbe diffraction limit of light conventional fluorescence microscopes are limited in their resolution to ~250 nm (approximately equal to half the wavelength of the used light beam). Nevertheless, cellular structures including synapses but especially the proteins they are composed of are far smaller, partially by more than a factor of 10. As a conventional fluorescent microscope illuminates all fluorophores within a spot at the same time, they also emit light at the same time. Therefore, two points in closer proximity than the full-width-half-maximum (FWHM) of their point spread function (PSF) will be difficult to resolve, as both PSF overlap to a certain degree. The invention of super-resolution microscopy broke the diffraction barrier within the last 20 years and opened a complete new field of science. In our lab we mainly use the super-resolution technique Stimulated Emission Depletion microscopy (STED).
For STED microscopy, two overlapping, synchronized laser beams are used, which arrive at the same sample position consecutively whereas the first laser excites the fluorophore (excitation laser) and forces an electron to a higher energy level whereas the second one (STED or depletion laser) de-excites/depletes it and forces the electron back to the ground state. In the region where both lasers are present, stimulated depletion is happening, while in a focal spot where just the excitations laser is present, spontaneous emission and therefore fluorescence is observed.
Electrophysiology
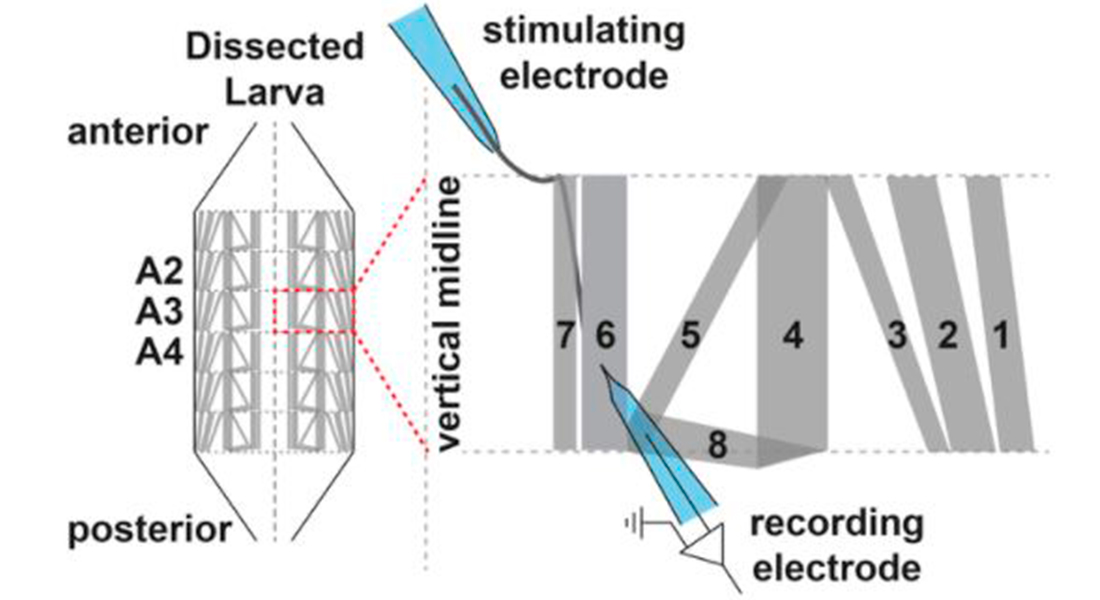
We use the Drosophila larval neuromuscular junction as a model to investigate neurotransmission. Here motoneurons innervate muscle cells forming a large number of synaptic connections. We insert sharp glass electrodes into larval muscles and measure the electrical properties of the membrane by making use of two different techniques:
Current clamp records changes in the membrane potential. Two electrode voltage clamp records the current injected to maintain a fixed holding potential of the muscle membrane. We investigate evoked neurotransmission by stimulating the presynaptic axon as well as spontaneous activity. We avail of different pharmacological agents to identify underlying mechanisms of synaptic function.
Computational Biology & Mathematical Modelling
Experimental Data is not always sufficient to fully understand the biological processes under investigation. Therefore, we employ computer-aided methods that help us build simulations and models of the synapse. Making assumptions about what processes are involved in a synaptic process, we formulate mathematical equations and have the computer solve them.
The simulations we derive from this can then be compared to experimental data. If our simulations do not agree with the experimental data, we know that we are making the wrong assumptions and can adapt the model until we find which underlying process we are missing.
Lab members
| Name | Title | Job responsibilities | |
|---|---|---|---|
| Search in Name | Search in Title | Search in Job responsibilities | |
| Alexander Matthias Walter | Professor | Walter Lab |
|
| André Rajoo | Laboratory Assistant | Walter Lab |
|
| Anna Linnea Müllenborn | Bachelor student | Walter Lab |
|
| Anna Schrøder Lassen | Research Assistant |
|
|
| Asger Marschall-Booth | Bachelor student | Walter Lab |
|
| Christian Fokdal Christensen | Postdoc | Walter Lab |
|
| Kavya Vinayan Pushpalatha | Postdoc | Walter lab |
|
| Keagan Scott Chronister | Research Assistant | Walter Lab |
|
| Maider Gonzalez Muxika | Laboratory Assistant | Walter Lab |
|
| Pontus Benjamin Scott Leblanc | Research Assistant | Walter Lab |
|
| Thiago Cordeiro Moulin | Guest Researcher | Walter Lab |
|
| Vera Kovaleva | Postdoc | Walter Lab |
|
